Releasing Biological Data from COVID-19 Lockdown
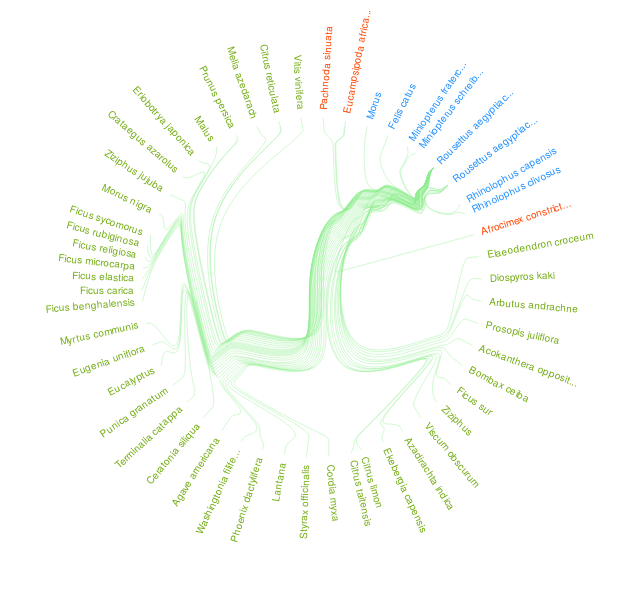
Figure 1. An interaction visualization of Rousettus aegyptiacus bat species indexed by GloBI on 2020-05-26 and made available by Quentin Groom's manually transcribed literature references via https://doi.org/10.5281/zenodo.3782323. Please visit a full list of associated references and associated visualization .
Wouldn’t it have been better if we had known more about bats before the COVID-19 crisis? One in five of all mammals are bats; there are over 1400 living species (Simmons and Cirranello, 2020). Bats live on every continent except Antarctica, and in every imaginable habitat. Yet we know so little about most species. Where do they live? What do they eat? What other organisms do they interact with? How does this all relate to the transmission of viruses between bats, and between bats and other organisms?
During the current lockdown, the museum collection community established an international COVID-19 Task Force (https://cetaf.org/covid19-taf-communities-taking-action). COVID-19 research touches upon many aspects of science and data management. SARS-CoV-2, the virus that causes COVID-19, is likely to have originated from bats, and knowledge about bat ecology and interactions could be critical to avoiding crises like this in the future (Andersen et al., 2020).
Our team specifically looked at the availability of data on bats, viruses, and other coronavirus hosts. While there is a lack of basic knowledge, the problem is larger than that: much of the knowledge that we do have is not readily accessible. It is tucked away in undigitized museums, in undigitised literature, in closed access publications, or it is hard to find due to poor data management. Yet, we do have the tools and infrastructure to make this knowledge accessible. There are collection digitization programs, such as ADBC in the USA and DiSSCo in Europe. The publisher Pensoft is pioneering ways to make interaction data digitally accessible from publications. And repositories such as Zenodo, GloBI and TreatmentBank provide a place and methods to make access and publication of data easier. So, in a short space of time our small, eclectic team of scientists was able to mobilise a surprising large amount of data on the subject. Currently, we have added 17,123 new distinct taxonomic names and 85,493 interactions to GloBI (Poelen et al., 2020).
My small contribution was mobilising species interaction data to GloBI, which, to be perfectly honest, is not that difficult. After conducting a literature search for the interactions of the Egyptian fruit bat (Rousettus aegyptiacus), I cloned a template repository from GloBI and created my own dataset on GitHub (https://github.com/qgroom/batinterations). I then learned how to get this validated and harvested by GloBI and for an archive copy to be stored on Zenodo (https://doi.org/10.5281/zenodo.3782323. Now, not only are these standardized data available to me, they are for everyone else too.
Alexander Graham Bell said that “when one door closes, another opens; but we often look so long and so regretfully upon the closed door that we do not see the one which has opened for us”. For all of us the COVID-19 crisis has seen many doors close, but despite the physical restrictions of lockdown, data are still just as mobile. One of the doors that is opening is the one to more open data that can contribute to a better understanding of the natural world and the interactions between different species in it. Let’s give that door a nudge.
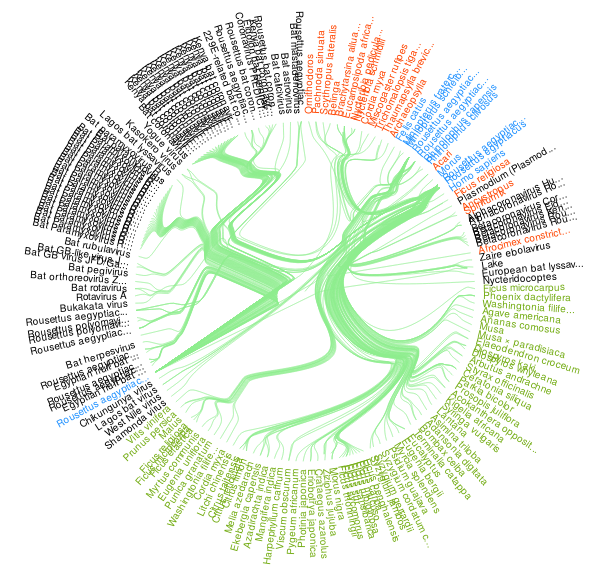
Figure 2. A visualization of all Rousettus aegyptiacus bat species interactions indexed by GloBI on 2020-05-26. Please visit a full list of associated references and associated visualization .
References
Andersen, K.G., Rambaut, A., Lipkin, W.I., et al. 2020. The proximal origin of SARS-CoV-2. Nat Med 26, 450–452 . doi:10.1038/s41591-020-0820-9.
Poelen, J.H., Upham, N.S., Agosti, D., et al. 2020. CETAF-DiSCCo/COVID19-TAF biodiversity-related knowledge hub working group: indexed biotic interactions and review summary (Version 0.2) [Data set]. Zenodo. doi:10.5281/zenodo.3839098 .
Simmons, N.B. and Cirranello, A.L. 2020. Bat Species of the World: A taxonomic and geographic database. https://batnames.org
Acknowledgements
Thanks to the Consortium of European Taxonomic Facilities (CETAF), the DiSSCo Infrastructure, iDigBio, Nancy Simmons (American Museum of Natural History), Donat Agosti of Plazi, whose goal it is to liberate biodiversity data from the dusty grave of, closed access and PDF-imprisoned literature, and by Jorrit Poelen who runs the Global Biotic Interactions database (GloBI).


