This page shows the current state of integrations between Plazi annotated publications, Zenodo and GloBI. This integration originated as part of developing a biodiversity-related knowledge hub on COVID-19 via COVID19-TAF - Communities Taking Action, a community-rooted initiative raised jointly by CETAF and DiSSCo.
These integration are very much a work in progress and will hopefully lead to an increased visibility of Pensoft publications that make claims about species interactions.
edit this page / edit collection list / ask a question / contribute data
events / integration status / integration profiles / translation tables / supported biotic interaction terms
Events
24 April 2020 - Exploratory Plazi<>Zenodo<>GloBI integration meeting with Jorrit (GloBI), Donat (Plazi), Marcus (Plazi) and Alex (Zenodo). meeting notes .
18 May 2020 - Continued Pensoft<>Plazi<>GloBI integration meeting with Jorrit, Lyubo, Mariya, Donat, Guido, Teodor, Georgi. meeting notes .
16/17 July 2020 - Guido adds Plazi-mediated publications to Zenodo using the custom “host of” Zenodo annotation that Alex implemented (e.g., Luo et al. 2018). Soon after, Jorrit enabled the indexing of these Zenodo pubs. For context, see related github issue and related tweets by Donat and GloBI .
2 Oct 2020 - Jorrit uses a newly added feature of Nomer to link taxonomic names indexed by GloBI to 41,894 Plazi treatments, 27,721 Plazi taxonomic concepts and direct links to 9,346 associated scientific publications. The methods and results of the link process are described in Poelen, Jorrit H. (2020). Global Biotic Interactions: Taxon Graph Patches (Version 0.5) [Data set]. Zenodo. http://doi.org/10.5281/zenodo.4062711 and Poelen, Jorrit H. (2020). Global Biotic Interactions: Taxon Graph (Version 0.3.26) [Data set]. Zenodo. http://doi.org/10.5281/zenodo.4062765 .
16 Nov 2020 - Katja, Marcus, Donat and Jorrit meet to discuss a semi-automated species interaction transcription workflow using existing GloBI<>Plazi<>BLR workflows. See #collaborative-transcription.
Integration Status
Click on badges to browse/download indexed records or inspect automated reviews.
| status | institution/collection | platform | contact |
|---|---|---|---|
| Plazi / Plazi Plazi-covid19 / Covid19 related literature list | google-sheets | Markus; Donat | |
| Plazi / Plazi plazi-zotero / Plazi annotated Zotero citation list | zotero | Markus; Donat | |
| Zenodo / Zenodo plazi-zenodo / Plazi annotated Zenodo publications | zenodo | Alex; Markus; Donat |
Integration Profiles
Integration profiles are descriptions on how data flows from one system to the next.
google-sheets
| Plazi Google Sheets | GloBI Integration Profile |
|---|---|
| authors | ? |
| actors | ? |
| integration method | Plazi team manually gathers literature on COVID-19 and extract virus-host annotations. These host-virus annotations are entered in a online spreadsheet. This Plazi curated spreadsheet is contiously tracked and indexed by GloBI. Now, GloBI helps make the literature annotations curated by the Plazi team more visible. |
| diagram | ? |
| examples | see related issue 490 |
Zotero
| Zotero | GloBI Integration Profile |
|---|---|
| authors | … |
| actors | … |
| integration method | Plazi team takes manually curated spreadsheets to automatically populate an annotated Zotero reference list of COVID-19 publications. Also, interested parties can append relevant publication to the Zotero reference list to allow to outside, non-Plazi contributions. The each contributed citation, Plazi help to extract and add relevant virus-host annotations and records them manually in the curated spreadsheet. |
| diagram | … |
| examples | see related issue 487 |
| status | method not implemented because alternatives exist (googlesheets, Zenodo) |
Zenodo
| Zenodo | GloBI Integration Profile |
|---|---|
| authors | Alex, Markus, Donat, Jorrit |
| actors | … |
| integration method | Plazi team uses each invidual reference in annotated Zotero citation list and creates an individual Zenodo publication. These Zenodo publications are annotated with relevant taxonomic, interactions, locality etc. properties. Interaction annotations are encoded in a key value scheme that allows for encoding interaction triples, taking inspiration from hexastore. For more information see Virus host relationship custom metadata: feature request. |
| diagram | … |
| examples | see related issue 488 |
Collaborative Transcription
| Collaborative Transcription | GloBI Integration Profile |
|---|---|
| status | under development |
| authors | Marcus, Katja, Donat, Jorrit |
| actors | … |
| integration method | … |
| diagram | 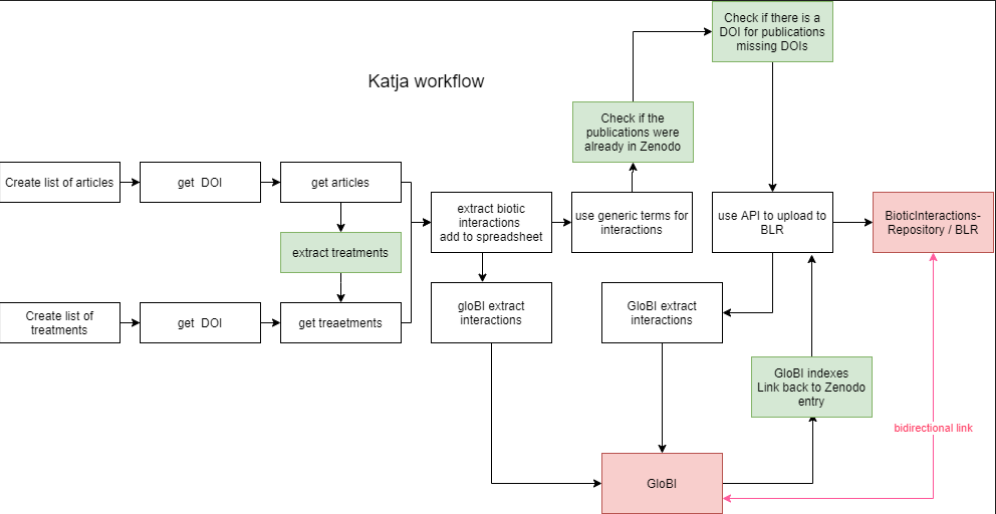 see also https://drive.google.com/file/d/1-lSC0-pqOYIUBDtij0_XDfSY7yBHnkhq/view?ts=5fb2db8a see also https://drive.google.com/file/d/1-lSC0-pqOYIUBDtij0_XDfSY7yBHnkhq/view?ts=5fb2db8a |
| examples | See https://github.com/globalbioticinteractions/globalbioticinteractions/issues/559 for example |
Plazi Transcription Provenance
| Plazi Transcription | GloBI Integration Profile |
|---|---|
| status | under development |
| authors | Jorrit |
| actors | … |
| integration method | Plazi facilitates scanning and extracting knowledge from existing taxonomic literature. This integration profile attempts to give a high level summary of the provenance of Plazi’s structured treatments. Roughly, after selecting a print media publication of interest, some entity (not plazi, but commercial folks or other specialists) scans (1) and OCRs (2) the results. Then, Plazi team uses their GoldenGATE Imagine tool (https://github.com/gsautter/GoldenGATE-Imagine) to extract the meaning of the text using a variety of techniques (e.g., templates) to annotate the meaning of the text (e.g., start of treatment, scientific name, location, bibliography). Then, their tool turns this annotated text into their own XML format (4), and stores them in the TreatmentBank XML corpus (5). This XML corpus lies at the foundation of all applications of Plazi’s treatment bank. Examples include, but are not limited to: publication to Biodiversity Literature Repository (BLR) on Zenodo, registration with Global Biodiversity Information Facility (GBIF, https://gbif.org), and Synospecies (https://synospecies.plazi.org/). |
| diagram | 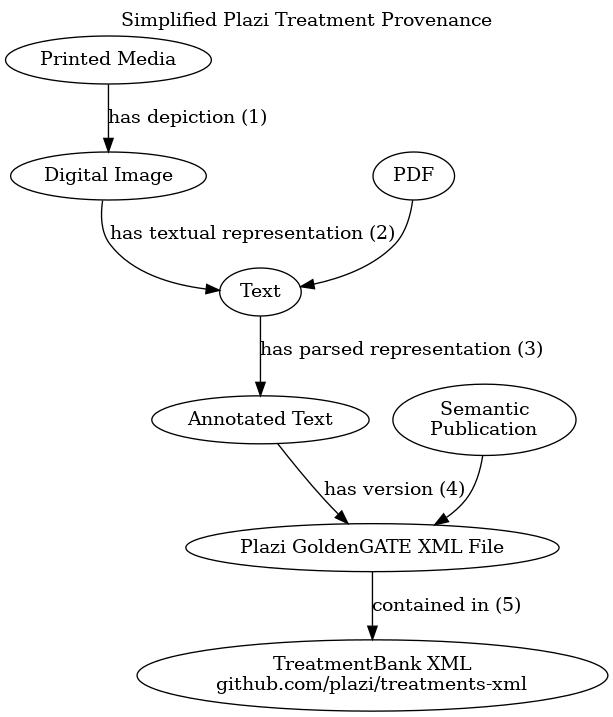 |
| examples | See e.g., https://github.com/globalbioticinteractions/globalbioticinteractions/issues/783 |
Interaction Types
Supported Terms
Translation Tables
GloBI uses a subset of biotic interaction (or association) terms defined in the OBO Relations Ontology (OBO RO) to help classify and index biotic associations in collection records. Verbatim association types (e.g., on, parasite of, found on) are explicitly mapped into these OBO RO terms using translation tables. GloBI keeps a default translation tables and specific collection may choose to provide their own (see e.g., INHS-Insects ).
| resource | description |
|---|---|
| OBO Relations Ontology project page | OBO RO contains many kinds of terms, not just biotic associations terms |
| List of OBO RO Biotic Interaction Terms with definitions | a table of RO biotic interaction terms and their definitions (if available) |
| List of GloBI Supported Interaction Terms | subset of RO interactions terms that GloBI uses for indexing |
| Default Verbatim Terms Translation Table | the translation table used by GloBI to maps verbatim interaction terms to supported interaction terms |
| Example of Custom Verbatim Terms Translation Table | if provided/needed, GloBI can use a custom mapping provided by a collection |
The OBO RO is far from complete and we expect to add new terms and improve definitions as needed. Also, GloBI translation tables can be easily updated when needed. Please open an issue if you have questions or suggestions.




